Characterization of transcriptomic and epitranscriptomic states between nuclear and cytoplasmic RNA using Nanopore-seq
Our lab uses Nanopore sequencing of cDNA and native RNA to explore transcriptomic and epitranscriptomic differences between nuclear and cytoplasmic compartments. We aim to uncover how RNA modifications and processing shape RNA fate and function across cellular contexts.

Exploring the human ribosome biogenesis landscape using NanoRibolyzer
In our lab, we developed a custom experimental and bioinformatic pipeline, NanoRibolyzer, to cluster and quantify ribosome biogenesis intermediates from long-read Nanopore data. Combined with our optimized nuclei isolation protocol, it enables spatial resolution of pre-rRNA and mature rRNA populations across nuclear and cytoplasmic compartments. This approach provides unprecedented insights into the dynamics of ribosome biogenesis, ribosomal RNA processing and modification.
Real-time transcriptomic profiling in distinct experimental conditions using NanopoReaTA
Real-time analysis tools have great potential for academic and clinical applications, as they allow for data acquisition and analysis to occur simultaneously during sequencing. Our group is interested in developing tools for real-time analysis using nanopore sequencing.
One of our latest developments is the development of NanopoReaTA, a tool that enables the detection of transcriptional changes between distinct conditions in real-time during sequencing (Wierczeiko et al. 2022). We are constantly exploring new avenues to enhance the capabilities of Nanoporeata and other tools to enable even more precise and powerful real-time analysis.

We showcase NanopoReaTA as a tool for real-time transcriptomic analysis across diverse settings. Its utility is demonstrated in profiling human cell populations, enriched transcripts, stress responses, and genetic perturbations in yeast.

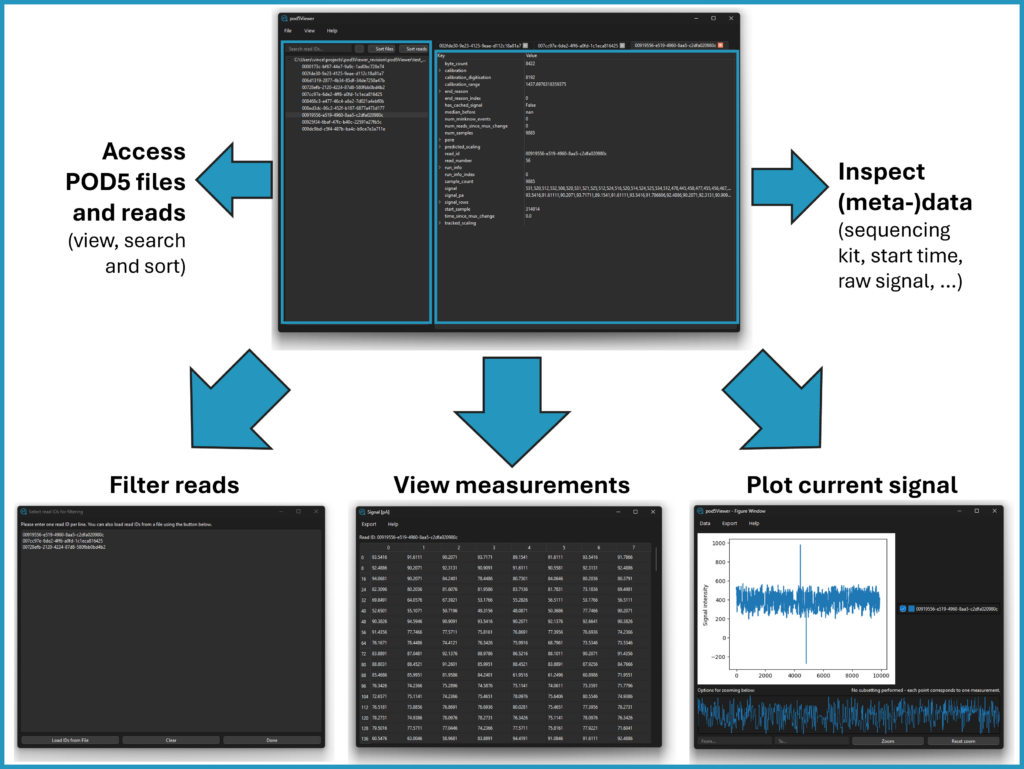
Pod5Viewer: making raw Nanopore data accessible
Our group contributed to the development of Pod5Viewer, a graphical interface (GUI) designed to easily explore raw Nanopore sequencing data in pod5 format